set.seed(2020)
# Load needed packages ----
suppressPackageStartupMessages(library(ggplot2))
suppressPackageStartupMessages(library(ggdist))
suppressPackageStartupMessages(library(raster))
suppressPackageStartupMessages(library(tidyverse))
# Load species and environmental data ----
sp <- c("lyva", "eclu", "trve")
pts <- lapply(sp, function(x){
read.csv(paste0("../data/", x, "/", x, "_filt.csv"))[,1:2]
})
# Load predictions ----
pred <- lapply(sp, function(x){
f <- list.files(paste0("../results/", x, "/predictions/"), full.names = T,
pattern = "current")
f <- f[grep("cont", f)]
f <- f[grep("mean", f)]
p <- raster(f)
# convert to 0 - 1 scale
p <- (p - minValue(p))/(maxValue(p) - minValue(p))
return(p)
})
# Now threshold to remove areas with very low ROR
for (i in 1:3) {
pred.vals <- raster::extract(pred[[i]], pts[[i]])
p10 <- ceiling(length(pred.vals) * 0.9)
thresh <- rev(sort(pred.vals))[p10]
pred[[i]][pred[[i]] < thresh] <- NA
}
# Sample 1000 points, using the ROR as a probability
spts <- lapply(pred, function(x){
rpts <- dismo::randomPoints(x, n = 1000, prob = T)
return(rpts)
})
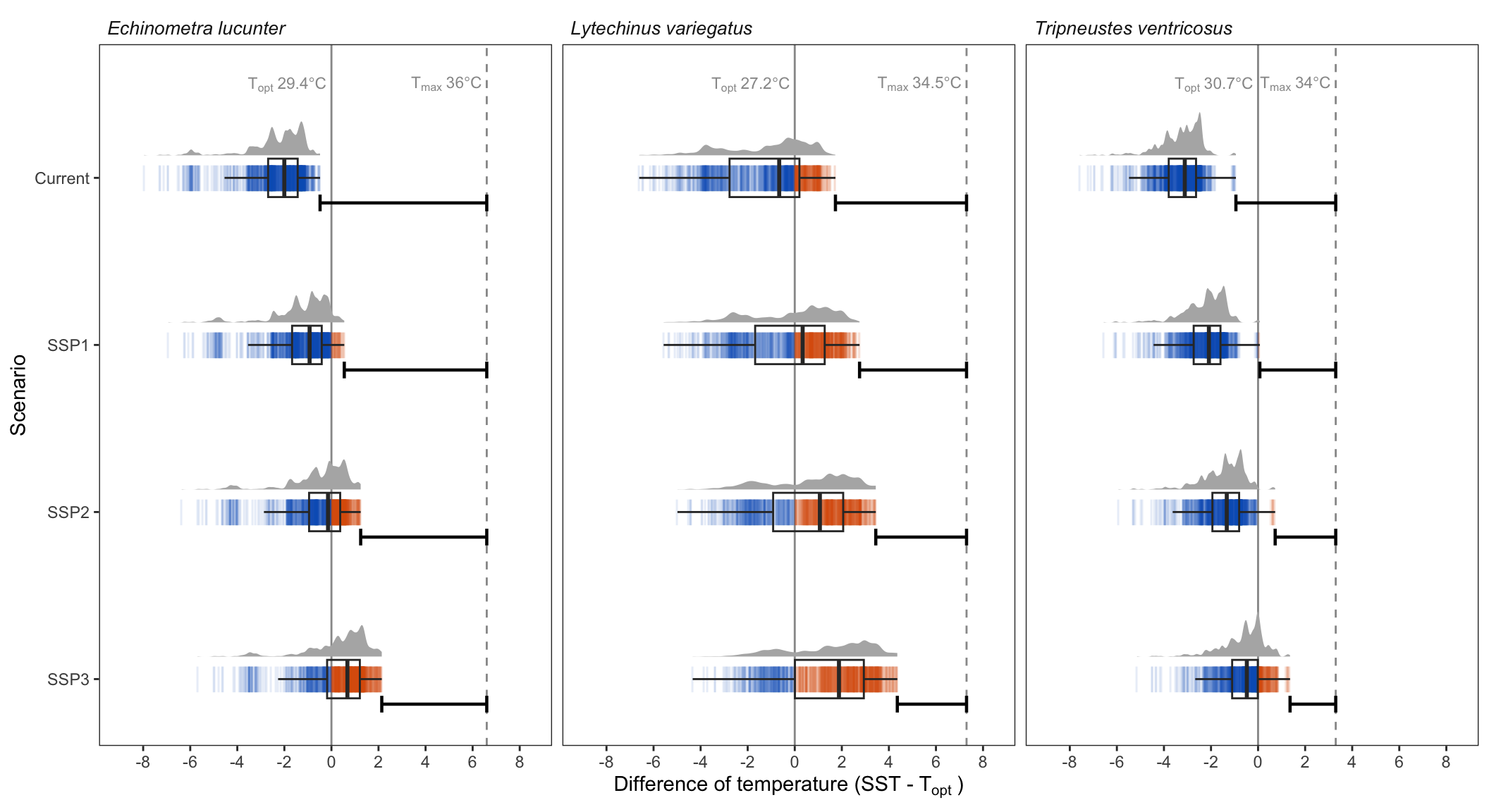
# Species temperature limits
lims <- data.frame(species = sp,
optimum = c(27.2, 29.4, 30.7),
upper_l = c(34.5, 36, 34),
lower_l = c(14.6, 14.3, 19.1))
lims$delta_lo <- lims$lower_l - lims$optimum
lims$delta_up <- lims$upper_l - lims$optimum
names(pts) <- c("lyva", "eclu", "trve")
curr <- raster("../data/env/crop_layers/BO21_tempmean_ss.tif")
ssp1 <- raster("../data/env/proj_layers/ssp126/BO21_tempmean_ss.tif")
ssp2 <- raster("../data/env/proj_layers/ssp245/BO21_tempmean_ss.tif")
ssp3 <- raster("../data/env/proj_layers/ssp370/BO21_tempmean_ss.tif")
baseproj <- raster("../data/env/ready_layers/sst_cur.tif")
curr <- projectRaster(curr, crs = crs(baseproj))
ssp1 <- projectRaster(ssp1, crs = crs(baseproj))
ssp2 <- projectRaster(ssp2, crs = crs(baseproj))
ssp3 <- projectRaster(ssp3, crs = crs(baseproj))
data <- data.frame(sst = 1, species = NA, scenario = NA)[-1,]
for (i in 1:3) {
c1 <- data.frame(sst = raster::extract(curr, spts[[i]]) - lims[i,2],
species = names(pts)[i], scenario = "current")
c2 <- data.frame(sst = raster::extract(ssp1, spts[[i]]) - lims[i,2],
species = names(pts)[i], scenario = "ssp1")
c3 <- data.frame(sst = raster::extract(ssp2, spts[[i]]) - lims[i,2],
species = names(pts)[i], scenario = "ssp2")
c4 <- data.frame(sst = raster::extract(ssp3, spts[[i]]) - lims[i,2],
species = names(pts)[i], scenario = "ssp3")
data <- rbind(data, c1, c2, c3, c4);rm(c1, c2, c3, c4)
}
# Classify values below/above optimum
data$sit <- ifelse(data$sst <= 0, "L", "H")
# Reorder scenarios
data$scenario <- as.factor(data$scenario)
data$scenario <- factor(data$scenario, levels = c("ssp3", "ssp2", "ssp1", "current"))
data.lims <- suppressMessages(data %>%
group_by(species, scenario) %>%
summarise(sst_max = max(sst), sst_min = min(sst)))
data.lims$up_lim <- rep(c(lims[lims[,1] == "eclu","delta_up"],
lims[lims[,1] == "lyva","delta_up"],
lims[lims[,1] == "trve","delta_up"]), each = 4)
data.lims$lo_lim <- rep(c(lims[lims[,1] == "eclu","delta_lo"],
lims[lims[,1] == "lyva","delta_lo"],
lims[lims[,1] == "trve","delta_lo"]), each = 4)
lims$labels <- paste0(expression(T[opt]), "~", lims$optimum, "*degree*C")
lims$labels_b <- paste0(expression(T[max]), "~", lims$upper_l, "*degree*C")
# Facet label names
supp.labs <- c("Echinometra lucunter",
"Lytechinus variegatus",
"Tripneustes ventricosus")
names(supp.labs) <- c("eclu", "lyva", "trve")
# Plot
p <- ggplot(data) +
geom_hline(yintercept = 0, linewidth = .5, color = "grey60")+
geom_hline(data = lims, aes(yintercept = upper_l - optimum),
linewidth = .5, color = "grey60", linetype = 2)+
ggdist::stat_halfeye(
aes(x = scenario, y = sst),
adjust = .5,
width = .3,
.width = 0,
justification = -.5,
fill = "grey70",
point_colour = NA
) +
geom_point(
# draw horizontal lines instead of points
# See: https://www.cedricscherer.com/
shape = "|",
size = 4.8,
alpha = .1,
aes(x = scenario, y = sst, color = sit)
) +
geom_boxplot(
aes(x = scenario, y = sst),
width = .23,
outlier.shape = NA,
fill = NA
) +
scale_color_manual(values = rev(c("#0B59BF", "#D65600")))+
scale_fill_manual(values = rev(c("#0B59BF", "#D65600")))+
geom_errorbar(data = data.lims,
aes(x = scenario, ymin = sst_max, ymax = up_lim),
linewidth = .8, color = "#030303", width = .1,
position = position_nudge(y = 0, x = -0.15))+
geom_text(data = lims,
aes(label = labels),
x = "current", y = -0.2, parse = T, vjust = -5,
size = 3, hjust = "right",
color = "grey60")+
geom_text(data = lims,
aes(label = labels_b, y = (delta_up - 0.2)),
x = "current", parse = T, vjust = -5.9,
size = 3, hjust = "right",
color = "grey60")+
theme_bw()+
ylab(expression("Difference of temperature (SST -"~T[opt]~")")) +
xlab("Scenario")+
scale_x_discrete(labels = c("SSP3", "SSP2", "SSP1", "Current"),
expand = expansion(add = c(0.4, 0.8)))+
scale_y_continuous(limits = c(-9, 8.5),
breaks = seq(-8, 8, by = 2))+
theme(panel.grid.major.y = element_blank(),
panel.grid = element_blank(),
strip.background = element_blank(),
strip.text = element_text(hjust = 0, size = 10, face = "italic"),
legend.position = "none")+
coord_flip()+facet_wrap(~species, labeller = labeller(species = supp.labs))
p